Why studying plankton ?
Plankton at the surface of the oceans includes a considerable diversity of bacterial, archaeal and unicellular eukaryotic cells along with their infecting viruses. Most of these microbial lineages have no representatives in culture collections and have yet to be properly placed in the tree of life. Our lab takes advantage of culture-independent DNA and RNA sequencing methods, largely through the prominent Tara Oceans projects, to explore the genomic diversity of plankton at large-scale. We combine state-of-the-art genome-resolved metagenomic and phylogenomic approaches to characterize the genomic content of the most abundant marine microbial lineages (from symbiotic bacteria to unknown eukaryotic species and entire new clades of viruses) and expose new branches in the tree of life relevant to our understanding of the ecology and evolution of plankton.
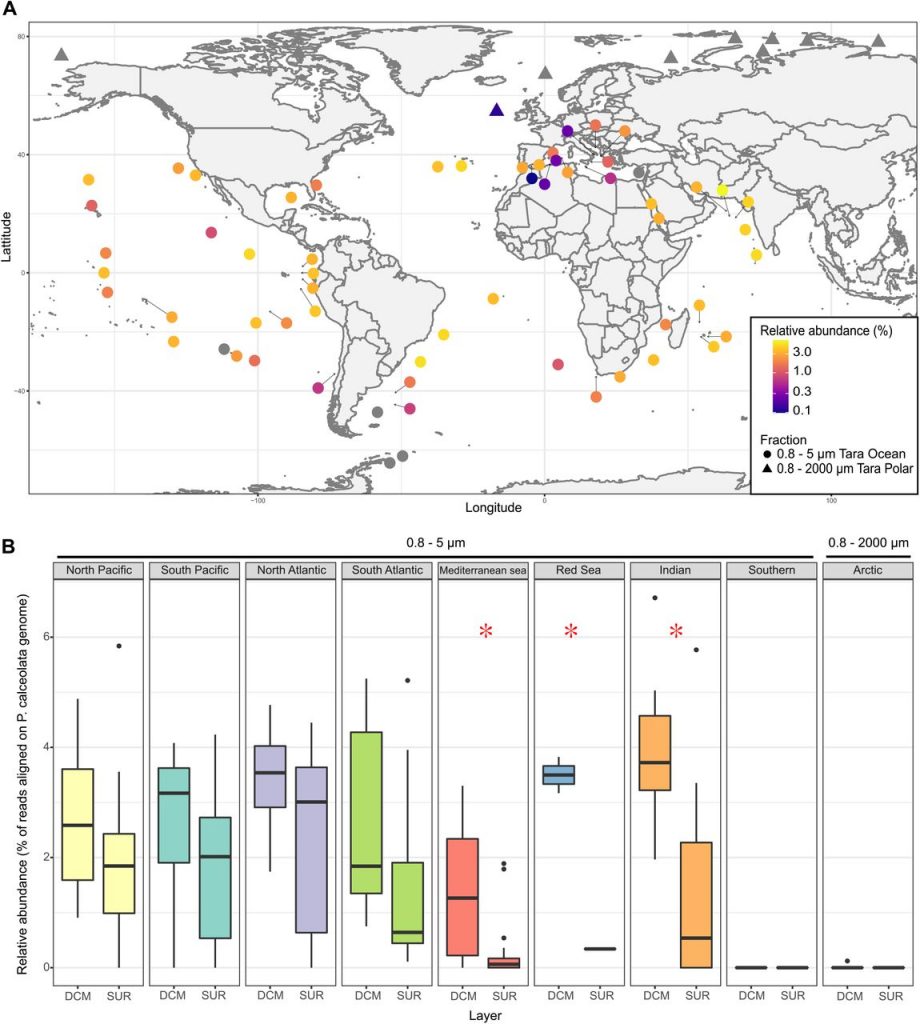
Omics : from sequence reads to communities
Our starting buiding bock is the sequence read. Either short (100 to 250 bp) or long (up to several hundreds of kb) reads that we obtain from the direct sequencing of DNA or RNA extracted from the sampled biomass. Assembly processes allows us to reconstruct pieces of genomes or transcriptomes that we cluster together to generate Metagenome-Assembled Genomes (MAGs) or MetaGenome-based Transcriptomes (MGTs) representing species-level genomic information. Combining these organism-level entities gives access to communities. We then predict the genes and annotate (identify their putative function) them, to understand and modellise their biological functions.
Functional insights
The microbial component of the ocean comprises more than 80% of the water column’ biomass. The oceanic microbiome deploys numerous interactions and survival strategies. But none of the species belonging to the ocean microbiome lives totally isolated from the others, on the contrary, these associations are crucial for the balance of ecosystems: either beneficial for both organisms or, on the contrary, deadly for the host, as in the case of parasitism, all levels of interaction are possible between these two extremes.
Today, our understanding of this microbial component is mostly based on a limited set of model organisms. Nevertheless, the enormous amount of environmental meta-“omics” data over the past years has deepened the knowledge on the “functional units” of genomes, with an increasing number of genes coding for novel proteins, or even those whose function is still unknown. Grasping the genomic basis of the biological and geochemical capabilities of the marine organisms will allow us to improve our understanding of the functioning of marine ecosystems. Exploring the adaptation of micro-eukaryotes to climate changes, unravel new genes’ function through experimental biology, seeking for markers of symbiotic and parasitic life-styles are some of the activities on our lab in quest of broadening our knowledge of the functional strategies deployed by the ocean microbiome.

Biogeography & Seascape Genomics

The study of the biogeography of organisms is a pathway to understanding the evolutionary processes they have undergone and their adaptive capacity. This understanding is crucial for planktonic organisms in the context of climate change. Indeed, it is essential to anticipate and understand the ecological and biogeochemical changes inherent to the restructuring of natural communities due to rapid climate change.
The context of the seascape* is a complication, but we want to exploit it to better reveal the links between genome dynamics and environmental variations. Among this information are parameters on adaptation and acclimation of organisms.
To understand these studies we need to implement comparative and evolutionary genomics approaches in natural populations, ecological or biogeochemical modeling.
