I’m a biologist specialized in bioinformatics and in the genomics of micro-eukaryotes. I completed my PhD in October 2014 at the Institut de biologie de l’ENS (Paris) in Eric Meyer’s laboratory studying molecular mechanisms of RNA interference pathways on the ciliate Paramecium tetraurelia. I switched to the environmental genomics domain to work on the transcriptomic activity of marine planktonic eukaryotes in the framework of Tara projects. My current research focus is on understanding the adaptation capacities of micro-eukaryotes to climate change.
email:
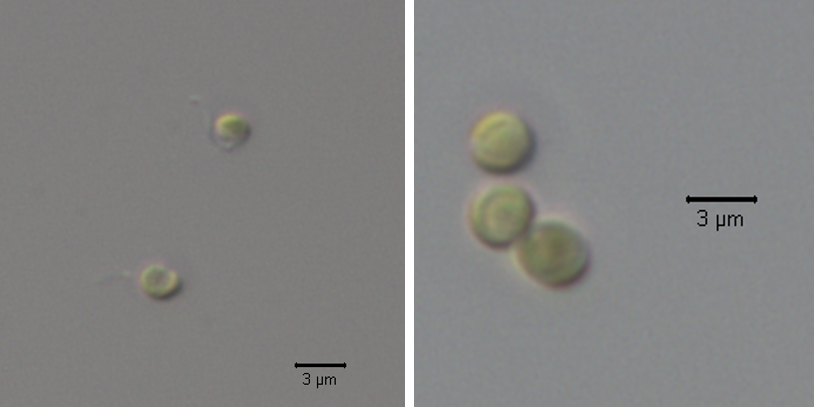
Acclimation of the picoeukaryote Pelagomonas to environmental changes
The smallest phytoplankton species are key actors in oceans biogeochemical cycling and their abundance and distribution are affected with global environmental changes. Picoeukaryotes (cells <3 µm) of the Pelagophyceae class encompass coastal species causative of harmful algal blooms while others are cosmopolitan and abundant in open ocean ecosystems. We use Pelagomonas calceolata as a model organism to study molecular mechanisms of acclimation and adaptation of open-ocean phytoplankton to climate change. The combination of in situ (Tara expeditions) and in vitro (cell culture) approaches is critical for providing an overview of the different scales of biological response to environmental change, from the molecular activity of specific genes to the environmental behaviour of wild populations across the world.
Other LAGE members involved : Nina Guérin, Adrien Thurotte, Chloé Seyman, Céline Orvain, Laurie Bertrand, Barbara Porro, LBGB team
Analysis of coral holobiont adaptation in the Pacific Ocean with -omics data

Increasing frequency and severity of marine heatwaves are causing global declines in coral reef ecosystems. Thermal adaptation and/or acclimatization capacities of coral holobionts are central for their resilience to these events. Because holobiont thermal response depends on multiple drivers, understanding the relative contributions of these factors to adaptive and/or plastic responses in the coral metaorganism is crucial for anticipating the impacts of global change on coral reefs. We analyzed DNA and RNA extracted from 800 coral colonies collected from 32 reef sites on 11 islands in the framework of the Tara Pacific expedition. These analyses provide a reference for the biological state of coral holobionts across the Pacific Ocean and highlight the importance of considering the different thermal acclimatization strategies of corals in order to better forecast coral reef persistence under continued ocean warming.
Other LAGE members involved : Mathieu Zallio, LBGB team
Past experiences
| 2016-now | Research Scientist (Genoscope, Laboratory of genomic analysis of eukaryotes) Project: Genomic and transcriptomic analysis of marine eukaryotes Research group: Dr. Wincker Patrick |
| 2015-2016 | Post-doctoral researcher (Genoscope, Laboratory of genomic analysis of eukaryotes) Project: Planktonic metatranscriptomes analysis from Tara Oceans samples Research group: Dr. Wincker Patrick |
| 2011-2014 | PhD thesis (UPMC, Paris-Saclay University, France) Project : Mechanisms and functions of the dsRNA-inducible RNAi pathway in Paramecium tetraurelia Director : Dr. Meyer Eric, team "Programmed genome rearrangements in ciliates" , IBENS |





